[EDIT: UMI-Tools open access publication is now out]
I’ve been working with Dr. Ian Sudbery (former CGAT fellow, currently Lecturer in Bioinformatics at the University of Sheffield) on a script called dedup_umi.py to correctly remove PCR duplicates from alignment files when using Unique Molecular Identifiers (UMIs). The script is available from GitHub as part of the UMI-tools repository. As part of the development I’ve been running some simulations to compare different methods which have proved quite illuminating. Before I go into the details though, some background.
PCR duplication artifacts
Many sequencing-based techniques rely upon an amplification step to increase the concentration of the library generated from the fragmented DNA prior to sequencing. Following alignment to the genome, it’s usually preferable to identify and remove PCR duplicates as there are inherent biases in the amplification step as some sequences become overrepresented in the finally library compared to their true abundance in the fragmented DNA. The brilliant Picard tool is commonly used to identify and remove these PCR duplicates using their genomic coordinates. Of course, it’s also possible that two sequence reads with the same genomic coordinates will have the originated from two independent fragments. However, where the alignment density is essentially random (as in DNA-Seq) this is unlikely, even more so if paired end sequencing has been performed. The human genome has ~3 x 109 bases and the insert size will vary so the combinations of possible genomic coordinates for the paired ends is so vast we don’t expect to sequence two fragments with the same genomic coordinates.
Conversely, in RNA-Seq, some cDNA sequences will be highly abundant and we are therefore much more likely to sequence multiple fragments with the exact same genomic coordinates. This problem is most acute where a high number of PCR cycles have been used to generate the sequencing library, for example where the concentration of starting material in low, such as in single cell RNA-Seq as this necessitates many PCR cycles.
Using unique molecular identifiers to correctly identify PCR duplicates
Unique molecular identifiers (UMIs; also called Random Molecular Tags (RMTs)) are random sequences of bases used to tag each molecule (fragment) prior to library amplification (see figure below and Kivioja et al (2012), thereby aiding in the identification of PCR duplicates. If two reads align to the same location and have the same UMI, it is highly likely that they are PCR duplicates originating from the same fragment prior to amplification. We can therefore collapse all the reads with the same genomic coordinates and UMI into a single representative read and obtain an accurate estimate of the relative concentration of the fragments in the original sample.
 This figure is adapted from Islam et al (2014)
This figure is adapted from Islam et al (2014)
The problem
The use of UMIs as described above would work perfectly if it were not for base-calling errors, which erroneously create sequencing reads with the same genomic coordinates and UMIs and that are identical except for the base at which the error occurred. Base-calling errors therefore inflate the apparent numbers of unique fragments sequenced (see below). An additional source of errors comes from the amplification itself which can introduce errors during the DNA duplication step.
The solution
Ian initially identified this issue in his own data and wrote a python script utilising pysam to parse an alignment BAM file and inspect the UMIs of reads with the same genomic coordinates. He reasoned that sequencing errors would produce UMIs which are a single edit distance away from another UMI. Therefore, by forming an adjacency matrix it should be possible to identify the sequencing errors and collapse the reads to the true number of unique UMIs present (see schematic below for a simple example). The first step is to identify connected components, where UMIs are represented by nodes and edges represent links between nodes which are a single edit distance apart. The counts of each node are also retained. Within each component, the most abundant node is selected and all nodes within a single edit distance are removed. This process is repeated using the second most abundant node and so on until all edges are removed and the number of nodes remaining is inferred to be the number of unique fragments sequenced. We refer to this method as the “adjacency” method.
It turns out this works pretty well, however, one can envisage situations where this may not correctly identify the true number of unique fragments. For example, an error during the PCR amplication may create an apparently distinct UMI from which sequencing errors could created further UMIs, all of which are actually from a single unique fragment (see schematic below). A similar situation could also arise if one particular position in the reads was more likely to contain an error (this is quite common in illumina sequencing). In this case, one would see lots of UMIs with a single edit distance due to an error at the common position and further errors may be present off of this UMI.
 In order to deal with these more complicated networks of connected UMIs, I developed the “directional_adjacency” method. I reasoned that we could resolve these networks by forming edges between nodes where the count of node A was >= 2 x node B. Now, each connected component is inferred to originate from a single unique fragment.
In order to deal with these more complicated networks of connected UMIs, I developed the “directional_adjacency” method. I reasoned that we could resolve these networks by forming edges between nodes where the count of node A was >= 2 x node B. Now, each connected component is inferred to originate from a single unique fragment.
In addition to these methods, we also wanted to try a modification of the “adjacency” method in which the whole connected component is inferred to originate from a single unique fragment (“cluster”).
This issue has been previously observed by Islam et al (2014) who proposed that UMIs with less than 1% of the average counts should be removed “to account for UMIs that stem from PCR-induced mutations or sequencing errors”. We implemented this method within our tool for comparison (“percentile”).
Finally, we implemented a “unique” method, where all unique UMIs were retained. Note, this appears to be the method which is most commonly used. The schematic below shows a comparison of the various methods (I’ve obviously rigged it to make directional adjacency look the best).
The proof
We’re taking two approaches to prove the value of improved detection of PCR duplicates using the adjacency methods implemented in dedup_umi.py. Firstly, we’ve conducted simulations to show they consistently estimate the true number of unique fragments more accurately. Secondly, we’ve applied dedup_umi.py to multiple single-cell RNA-Seq studies and show an improvement, with respect to the distribution of UMIs, the quantification of spike-in RNAs and the clustering of cells. I’ll focus on the simulation for now and leave the single-cell RNA-Seq to another post when we’ve submitted the manuscript.
In order to compare the various methods, we wanted to know how the methods fared across a range of UMI lengths and depths of sequencing. For this I turned to python to replicate the process of amplification and sequencing in silico and model the impact of the various parameters on the efficacy of the methods. The whole process is contained in a standalone ipython notebook which you can view here. If you would like to download and run the notebook yourself you can do so from here.
In brief, the simulation starts with a set number of unique molecules (fragments), our “ground truth”. A UMI is then randomly assigned to each molecule and the pool of molecules amplified by duplicating each molecules for every in silico PCR cycle. The final pool of molecules are then “sequenced” to produce the reads. Within this process, base-calling errors are incorporated during the sequencing and substitutions are incorporated during the PCR cycles. Furthermore, amplification efficiency of each UMI varies to naively model the impact of amplification biases. Various methods are then applied to the reads generated to simulate the de-duplication process within dedup_umi.py and an estimate of the number of unique molecules is obtained. To compare the methods I used 2 metrics:
- The difference between this estimate and the ground truth
- The variability of the estimate for which I used the coefficient of variation (CV; sd/mean)
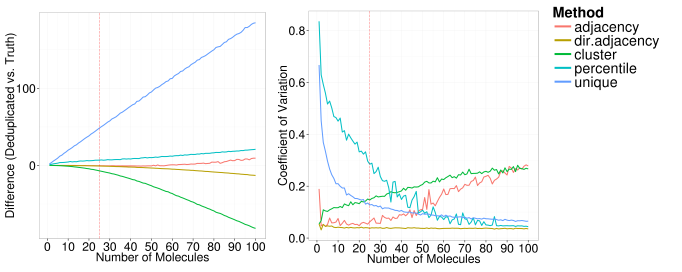
Each of the following plots below show the impact of changing one of the simulation parameters on theses two metrics. In each plot, the red dashed line shows the parameter value that was used in the other simulations.
We can see that under the assumptions of the simulations, the directional adjacency method is the least sensitive to variations in the input parameters.
The unique method (the method used in most publications) severely overestimates the number of unique molecules since each sequencing error or PCR error erroneously increases the estimate. As the number of starting molecules increase, all methods eventually struggle to identify the true number of unique molecules, with the adjacency, directional adjacency and percentile methods showing the best performance. As we would expect, the percentile method tends to overestimate the number of unique molecules since it fails to remove sequencing errors unless they are very rare events. The adjacency method tends towards an overestimation too, as it can only identify erroneous UMIs within a single edit distance. Interestingly, the directional adjacency method tends towards an underestimation as it can link together a string of UMIs each separated by a single edit distance. The directional aspect of the network is designed to avoid this happening between truly unique UMIs, however occasionally these will be collapsed together. The cluster method tends towards a more severe underestimation as it does not take counts into account at all and collapses whole networks into a single UMI.
Overall, the directional adjacency method shows very little sensitivity to the number of starting molecules and outperforms all other methods. As expected, the percentile method becomes more consistent as the number of molecules increase, since the variability between the counts in each one of the individual iterations is reduced. In contrast, the cluster and adjacency methods become less consistent as the number of starting molecules increases, as the variability in the topology of the networks formed will also increase.
As the UMI length increases (from a initial very short 3bp), the methods which utilise a network quickly improve in terms of accuracy and variability. However, the percentile and unique methods actually become less accurate as a longer UMI obviously means more probability of errors accumulating. Perhaps counter intuitively then, if you’re not going to account for errors when using UMIs, you’re better off not using a very long UMI.
As the PCR cycles are increased, all methods except directional adjacency show a decrease in accuracy and an increase in variability, with adjacency being the second most robust to high PCR cycle numbers. We see the same as we increase the DNA polymerase error rate. This confirms that the directional adjacency method is better able to handle errors originating from the amplification stage.
Is a non-heuristic solution possible?
The methods we propose are definitely an improvement on “unique” deduplication using UMIs within these simulations. We’ve also shown that the application of our proposed methods can have a real impact on biological interpretation of single cell RNA-Seq data (more to come on that soon). However, all implemented methods are heuristic approaches to the problem. My initial expectation was that we would be able to apply some probabilistic approach to determine the most likely set of UMIs which would account for the UMIs observed at a given genomic location. So far, we’ve been unable to develop such a method. If anyone would like to discuss alternative probabilistic approaches, or the script in general, please get in touch.







Reblogged this on Pythonic Biologist.
LikeLike
very interesting job. I am also working on UMI dedup now.
Have you think about UMI composition bias? In the data I have analysed there are more A or T enriched UMI. Is it caused by manufacture or just sequencing artifact? The data is produced by CEL-seq MARS-seq and sequenced using NEXTseq.
LikeLike
Hi there. We have looked at at composition bias, this is why our simulations sample the null from the UMIs sequenced, rather than using a uniform null. In general, I do see a bias: the UMIs definately do no fit an assumpution of uniform usage. In many cases there is a GC bias, but this GC bias seems to vary from library to library in a way we having been able to disentangle.
LikeLike
[…] to return to the original problem with our script. For a background on our UMI-tools see (CGAT blog – UMIs). The dedup_umi.py script in question takes the alignment file (SAM/BAM format) and returns an […]
LikeLike
Would you be able to provide me with some run time estimates for umi_tools dedup directional adjacency function? How long does it take to run the function on a bam with x mil reads paired-end using y cores and z GB RAM?
LikeLike
Hi Roy. To quote from the pre-print, “Time requirements for running dedup depend on number of input reads, length of UMI and level of duplication. Memory requirements depend on the number of output reads. On a desktop with a Xeon E3-1246 CPU, it takes ~220 seconds and ~100Mb RAM to process a 32 million read single-end input file with 5bp UMIs to ~700,000 unique alignments. Inputs with longer UMIs may take significantly longer”. Building the networks requires an all-vs-all comparison between the UMIs. The code for this has been sped up a little – see Speeding up python – but this step can could be time-consuming for alignment files with very high coverage at some positions.
LikeLike
[…] manner. For background on the network methods, please see our publication or my previous blog post. For the next major UMI-tools release (version 0.5), we’re planning to expand our support […]
LikeLike
[…] Source: Unique Molecular Identifiers – the problem, the solution and the proof | CGAT […]
LikeLike
Hi there,
I am about to use UMI-tools on some targeted DNA sequencing data and this post was of great help. Just wanted to let you know 🙂
LikeLike
Hi there, is there any UMI experts who could tell me how many different UMIs I could have for a 10bp oligo, where there are at least 2 differences between each UMI?
LikeLike
[…] methods to build and resolve networks of connected UMIs had also been incorporated (discussed in: Unique Molecular Identifiers – the problem, the solution and the proof). I think it would still be fair to say that UMI-tools was still “doing one thing well” […]
LikeLike